In biologics drug discovery, a biomolecule from its origins to the candidate drug status undergoes a variety of characterizations. Such characterizations are aimed to define the effectiveness, specificity, potency, adverse effects, pharmacology, and in vivo efficacy of the candidate biomolecules. Decades of advancements in biophysical methods that can analyze drug interactions with a particular target molecule have enabled these characterizations to be carried out without the need for reporter-based assays (or label-dependent assays).
Enabled by devices known as biosensors, the label-free methods can be used as matrix independent screening tools of candidate molecules against a target molecule of interest to determine interaction kinetics and thermodynamics in real-time. This blog highlights the synergy between label-free biosensors and the PipeBio cloud based bioinformatics platform - uncovering deeper insights into biological realities.
Introduction to label-free technologies
As the pharmaceutical industry became more profitable and globalized in the 1980s, the concept of innovation in drug discovery pipelines became decentralized into various disciplines: target identification & validation, lead identification & screening, lead optimization, upstream & downstream process development, manufacturing & quality control, and delivery optimization for clinical studies. Increased profit margins and curiosity in Biopharma attracted scientists with new ideas and investors with a mindset to increase the pace of drug discovery from laboratory bench to bedside treatment. This mindset drove the development of combinatorial chemistry and high throughput screening (HTS) methodologies. Candidate screening that was limited to a subset of label-dependent assays exploiting radioactivity, luminescence, or fluorescence became an obvious bottleneck due to inefficiencies stemmed from the labelling, lack of sensitivity, and cost.1 It became evident also that many of the above silos were unaware of the fate of the molecules or the impact of their characterizations as those molecules triaged from one discipline to another in a particular drug discovery pipeline.
Around the same time, a group of scientists and instrument developers decided to focus on the Surface Plasmon Resonance (SPR) based biosensors to characterize an interaction between a target and the candidate.2,3,4 Their desires lead to the studies of real-time interactions between an immobilized target molecule and the analyte molecules in solution without relying on a reporter molecule (label), unveiling the term ‘label-free’. The label-free SPR approach was originally envisioned as a method to determine affinity kinetics (based on the association and dissociation rates of binding), subsequent advances in optical and acoustic biosensing lead to technologies such as Bio Layer Interferometry (BLI)4,5, Surface Plasmon Resonance Imaging (SPRi)4, and Grating Coupled Interferometry (GCI)4, expanding the utility of label-free platforms (hence biosensors) for the analysis of biopharmaceuticals across various disciplines in a biologics drug discovery pipeline.
Label-Free Screening Platforms
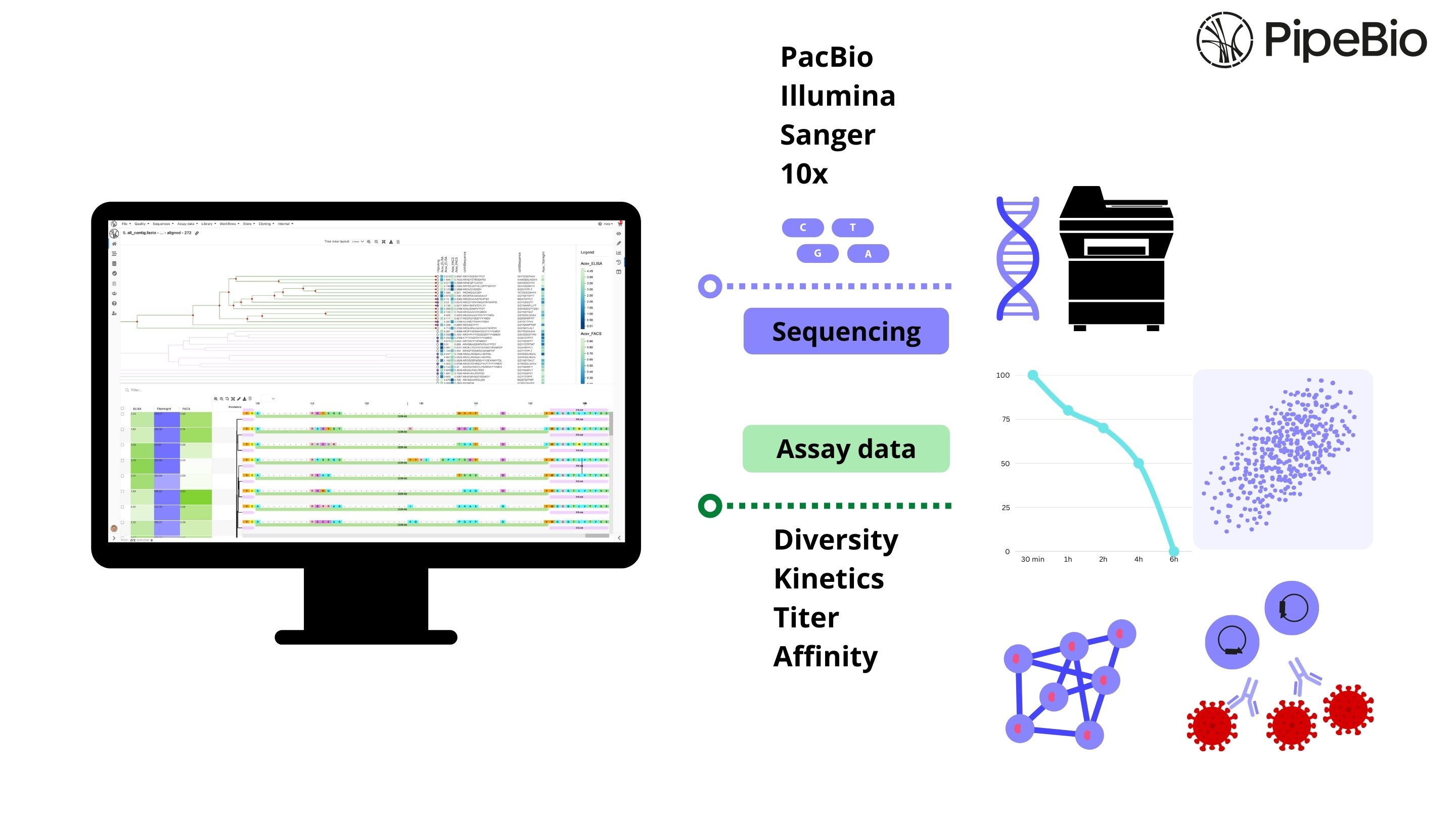
Cytiva Biacore-SPR
Biacore biosensor uses a chip surface against a glass prism while different types of flow cells are applied to the opposite side comprising a functionalized hydrogel. The mass increases when a molecule in solution (analyte) binds to a molecule immobilized to the surface (ligand) of a thin gold film. Changes to mass causes the refractive index of the solution near the film’s surface to alter, allowing an optically sensitive detection of a biomolecular interaction.
Sartorius/Fortébio Octet-BLI
Octet is a Bio Layer Interferometry (BLI) platform developed by Fortébio. BLI platforms are based on the “dip and read” fluidics-free and easy to use assay format. BLI systems measure the difference in reflected light wavelength between bound molecules and a reference surface. In addition to real-time kinetics and diversity assessments, the Octet BLI platforms measure titer and impurities in a high throughput manner.
%20platform%20developed%20by%20Forte%CC%81bio.jpeg)
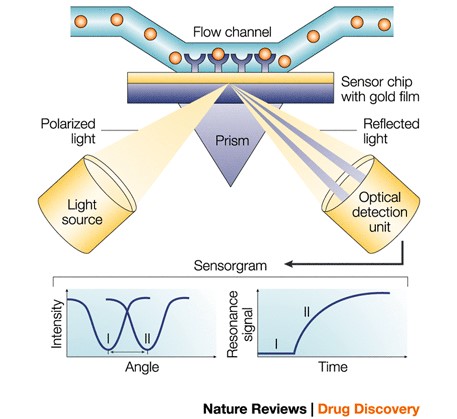
Carterra LSA HT-SPR
The Carterra LSA utilizes Surface Plasmon Resonance imaging (SPRi) to monitor binding interactions in real-time for up to 384 samples in parallel (SPR array detection). The LSA integrates flow printing technology via the 96-channel printhead for array building, while a large single flow cell (SFC) is used for chip surface preparation, regeneration, and pulse-free sample delivery for high-throughput kinetics. The LSA fluidics modes interchange between the 96 Printhead and SFC as defined by the specific protocol.
Figure: www.carterra-bio.com
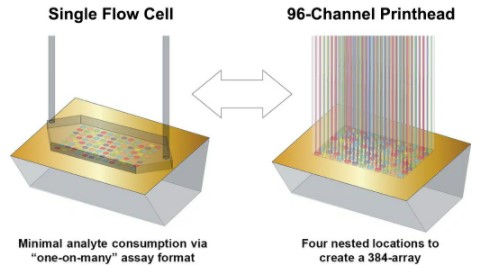
Bruker Sierra-SPR
The Sierra SPR-32 offers SPR+, a sensitive method of Surface Plasmon resonance imaging (SPRi) detection. SPR+ is capable of imaging two-dimensional arrays with a level of detection sensitivity not typically seen by the linear arrays. The optics unit of SPR+ combines traditional SPR imaging with high-intensity laser light and high-speed optical scanning. The system uses high-speed cameras to permit more response measurements per scan yielding a lower signal to noise and higher precision.
Figure: www.bruker.com
Gatorbio-BLI
Gator biosensor is a thin glass-rod consist of optical layers and an optimized surface chemistry incorporated to its business-end. Gator measures the interference pattern of white light reflected from an internal reference layer and a layer of immobilized biomolecules on the surface. Shifts in interference due to the accumulation of biomolecules are monitored in real-time to measure the rates of binding.
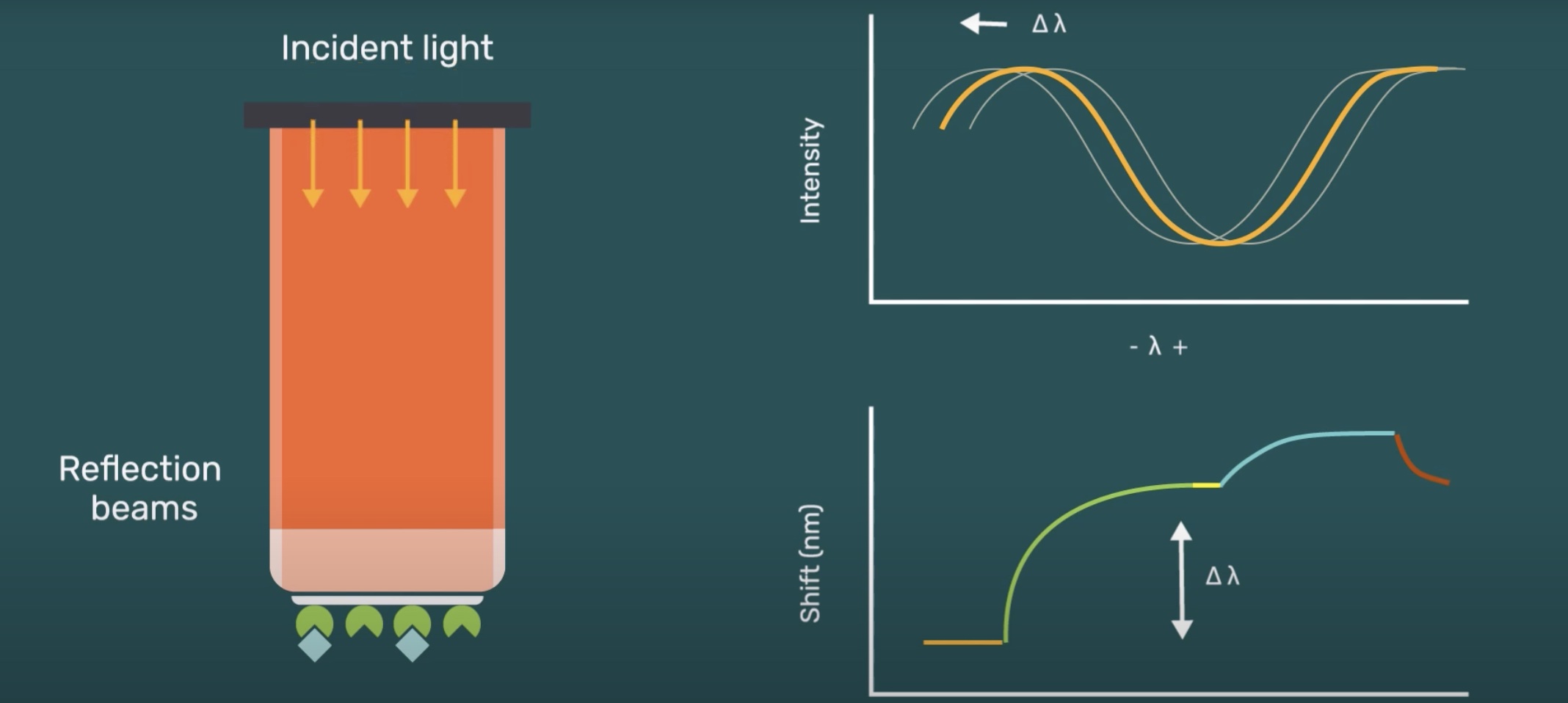
Creoptix
Creoptix™ WAVEsystem uses a unique Grating-Coupled Interferometry (GCI) approach, a method based on an improved waveguide interferometry, to monitor biomolecular interaction rate constants and concentrations of the interacting molecules. Like SPR, Waveguide Interferometry detects refractive index changes within an evanescent field near a sensor surface caused by the changes in mass during an interaction. The light in waveguide interferometry can travel over the entire interaction length differentiating it from SPR. However, the evanescent field does not penetrate as deep into the sample/matrix minimizing the bulk refractive index changes typically caused by temperature or mechanical distortions.
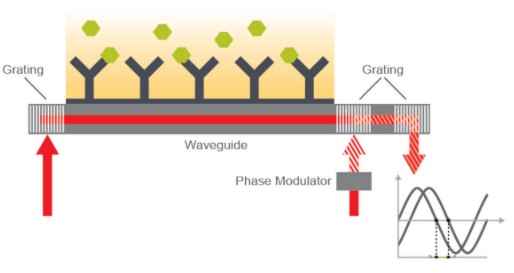
Nicoya OpenSPR
OpenSPR integrates digital microfluidics (DMF), artificial intelligence (AI), and nanotechnology to provide label-free interaction analysis. Instead of a thin gold film for biosensing, the OpenSPR platform uses gold nanoparticles to detect biomolecular interactions. Disposable fluidics components pertaining to DMF technology adopts low-volume crude samples and makes no demand for fluidic maintenance.
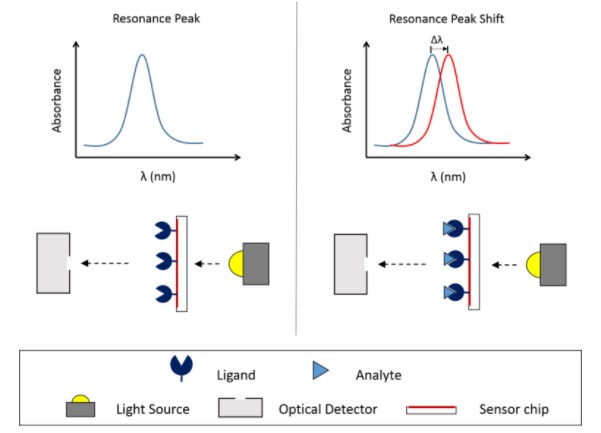
XanTec Bioanalytics GmbH
XanTec's surface plasmon resonance (SPR) platform measures binding kinetics, thermodynamics, and concentration of biomolecules in complex media. This platform can also quantify adsorption and desorption processes in non-biological systems or to follow the course of solid phase-based chemical reactions on the chip surface using organic solvents matrices.
Malvern Panalytical
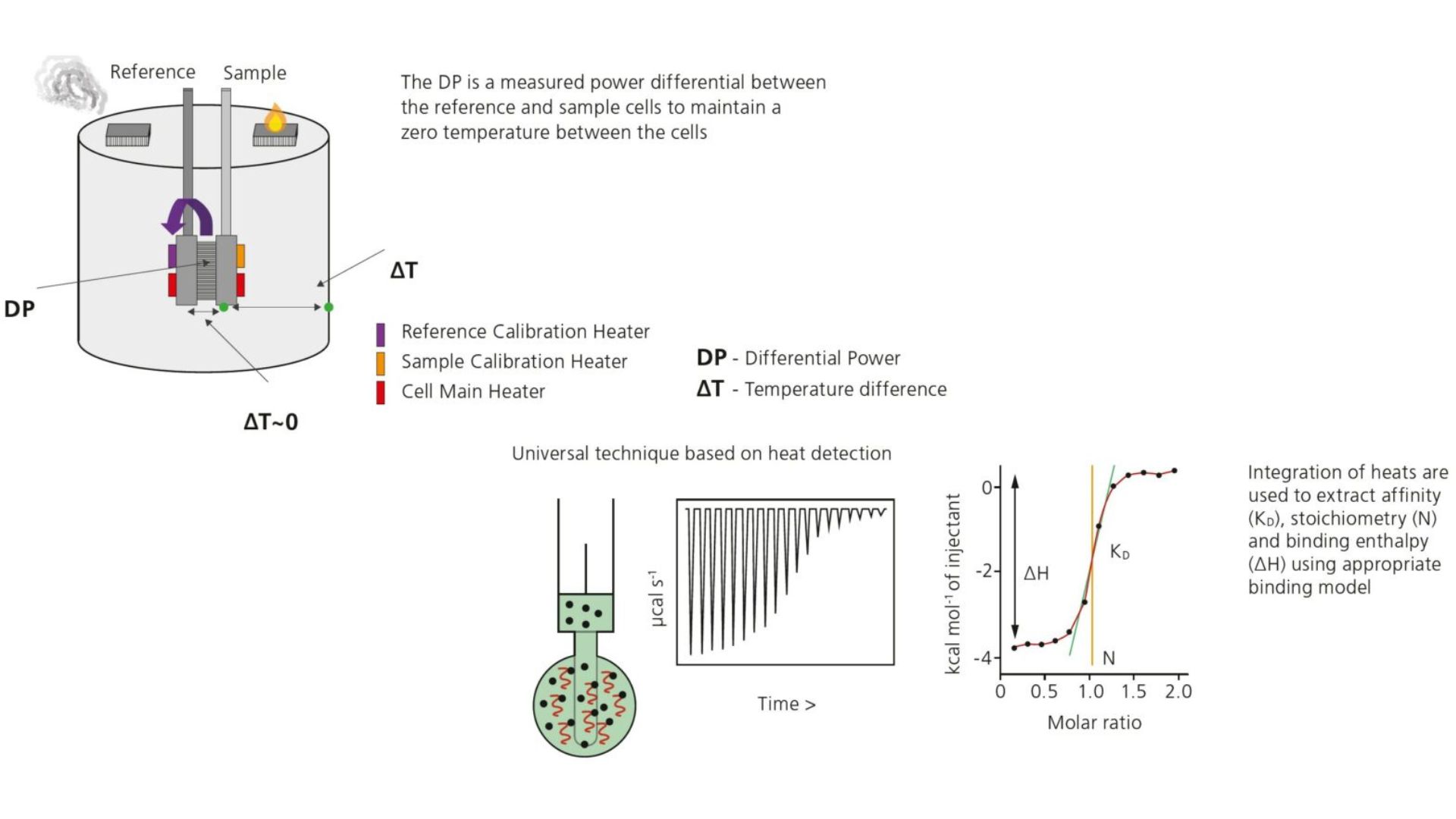
Isothermal Titration Calorimetry (ITC) is a label-free method for characterizing the interaction of molecules that either release or absorb heat upon binding. MicroCal ITC allows direct measurement of thermodynamic parameters of biomolecular interactions, namely the affinity (KA), enthalpy (ΔH), entropy (ΔS), and stoichiometry (n) in a single experiment. Thermodynamically favorable interactions have negative free energy values, ΔG = -RTlnKA = RTlnKD. The microcalorimeter has two cells: one is the Reference cell – usually contains water. The Sample cell contains one of the binding partners into which the other molecule is titrated using an injection syringe. Heat sensing devices detect temperature differences between the cells due to binding in the Sample cell and hence provide feedback to the heaters, which then compensate for the heat difference and return the cells to equal temperature.
Label-Free applications in Biologics Drug Discovery
With more than 60 market approvals and at least 500 more in development, Monoclonal antibodies (mAbs) remain as the richest macromolecular space for biologics drug discovery. Candidate antibodies and/or antibody fragments are pulled from large libraries derived from immunized animals, phage display libraries, or from antibody producing B cells leading up to thousands of mAb candidates. Characterization of large panels of mAbs can be time consuming and expensive. The advancements in label-free biosensors have enabled measurements directly in crude sample matrices such as cell lysates, serum, and growth media - extending the range of applications from the initial characterization of primary hits through pre-clinical profiling of therapeutically relevant lead mAbs. Label-free characterizations of mAbs involve real-time kinetics measurements, concentration determinations, diversity assessments, and mapping applications.4,6,7 Integration of robotics8 and advanced liquid handling9 options paved the way for label-free screening in high-throughput mode.
PipeBio complementarity with Label-Free Technologies for data-driven decision making in real-time
While the demand for therapeutic antibodies continue to grow, including therapies for infectious diseases such as COVID-19, researchers feel the demand to find the lead candidates rapidly and accurately. PipeBio, the cloud based bioinformatics platform combines a sequence database with a powerful analytical engine and a user-friendly interface to annotate high-volumes of data generated by Sanger and/or next generation sequencing (NGS) methods.
PipeBio enables an integrated platform for seamless import of information-rich data obtained from label-free interaction analyses. Multidisciplinary data interpretation and display options featured on PipeBio caters to a longstanding desire of wet-lab Scientists, i.e. to implement an integrated platform for data-driven understanding of quality attributes pertaining to their biologics derived from various disciplines within the antibody engineering and development pipeline.