With the increased throughput of single cell platforms and sequencing of immune repertoires, new challenges arise and the ability to map individual antibodies to its antigen is becoming increasingly important.
What is LIBRA-seq?
Mapping B cell receptor-antigen interaction at single cell level
Dr. Georgiev’s group at the Vanderbilt University has developed a high-throughput, highly-multiplexed sequencing-based technique that deciphers B cell receptor-antigen interaction at single cell level, called LIBRA-seq (Linking B cell Receptor to Antigen Specificity through Sequencing)3. The core of this technology is the DNA barcode conjugated and fluorophore tagged antigen. Such antigen can specifically bind to B cells expressing the corresponding BCR and then can be sorted based on the fluorescence and captured in droplets (one cell in one droplet) using the 10x genomics platform (Fig 2). The antigen barcode oligos can be read by sequencing together with its respective BCR sequence, simultaneously mapping antibody-antigen interaction at single cell level. The beauty of single cell sequencing is not only mapping antibody-antigen interactions, but also providing full-length paired heavy- and light-chain antibody sequences, which can be easily utilized to make recombinant antibodies.
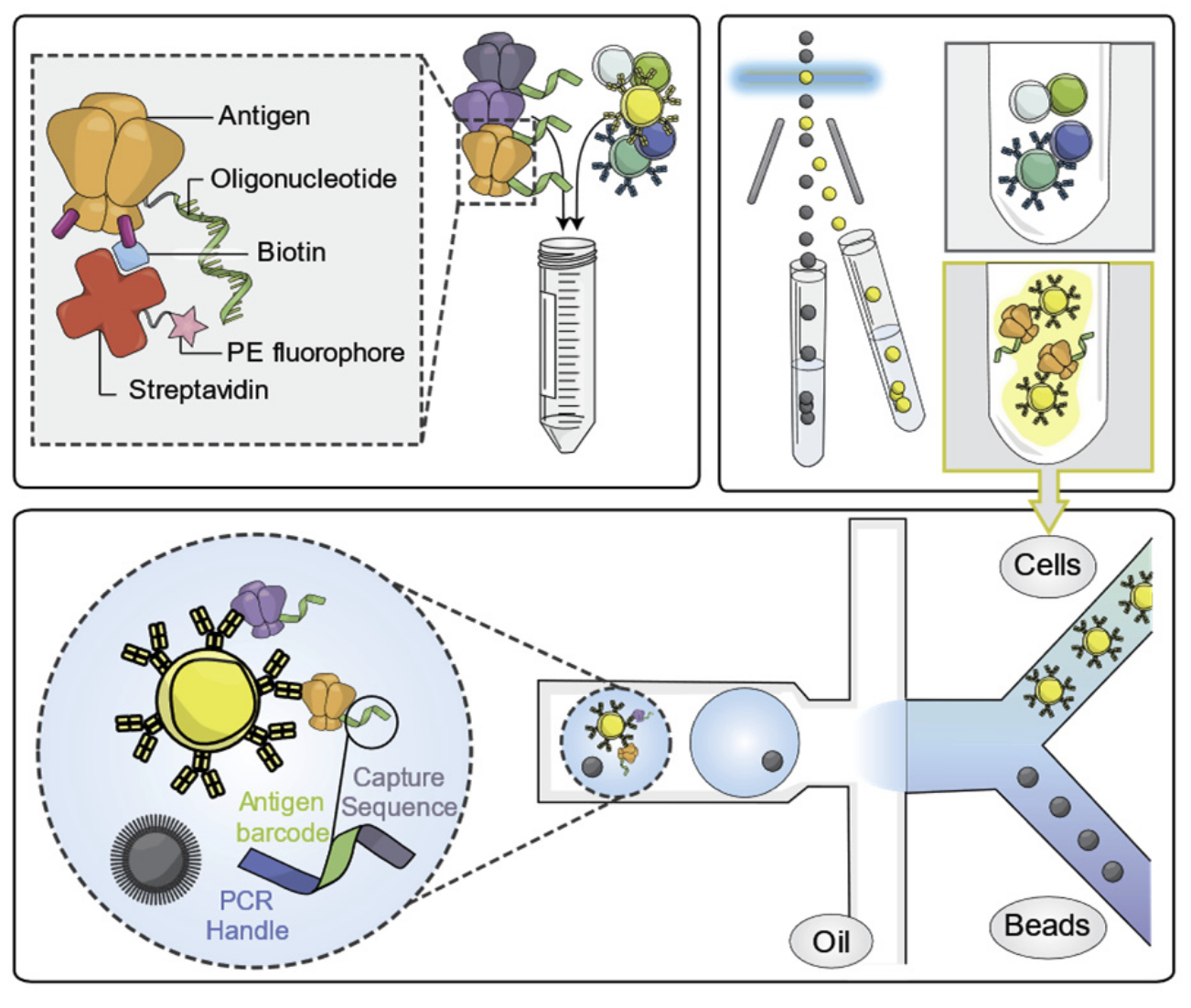
Briefly, LIBRA-seq contains 5 steps (Fig. 2)
Step 1: Mix B cells with DNA barcoded and fluorophore tagged antigens
Step 2: Sort antigen-bound (fluorescence positive) B cells by flow cytometry
Step 3: Single cell capture/profiling using 10X genomics platform
Step 4: Paired full-length heavy- and light-chain sequencing
Step 5: Bioinformatics analysis (recover BCR sequence and calculate antigen binding score)
The researchers validated LIBRA-seq using two engineered Ramo B cell lines expressing distinct BCR that neutralizes previously known antigens (e.g. VRC01 antibody neutralizing HIV-1 and Fe53 antibody neutralizing influenza). No cells with cross-reactivity to HIV-1 and influenza antigen were observed, suggesting a high specificity and sensitivity for LIBRA-seq (3).
Application 1: Identify a novel broadly neutralizing HIV-1 antibody 3602-870
To identify broadly neutralizing HIV-1 antibodies, the researchers performed LIBRA-seq by mixing the following antigens and B cells. Besides HIV-1 antigens, they also incorporated influenza antigens to detect the cross-reactivity of the antibodies. Antigen library: 5 trimeric HIV-1 Env proteins from HIV-1 strains with diverse clades + 4 trimeric HA proteins from influenza viruses
B cell resource: Blood sample from patient with chronic HIV-1 infection
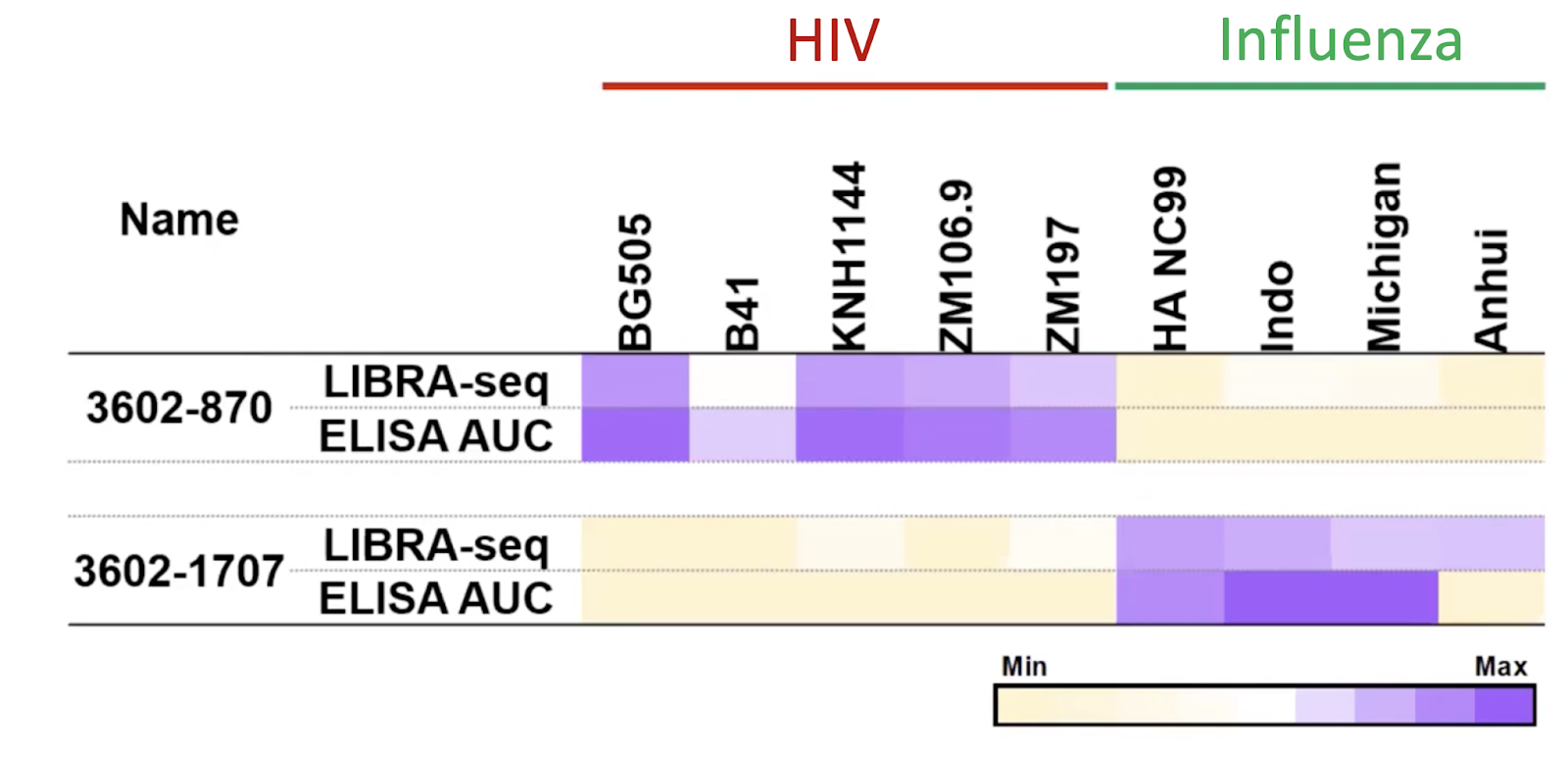
Using LIBRA-seq, the researchers count the number of unique antigen barcodes for each cell with the respective BCR sequence and compute a LIBRA-seq score to reflect the antigen specificity for each BCR. They prioritized the LIBRA-seq score and identified a novel broadly neutralizing HIV-1 antibody 3602-870 that targets all 5 HIV-1 antigens but not influenza antigens.
They further validated this antibody by ELISA, a well-established biochemical method to test antibody-antigen interaction. They found that the LIBRA-seq score is highly correlated with the ELISA results, indicative of robustness of LIBRA-seq (Fig 3).
Application 2: Identify ultra-potent neutralizing antibodies against SARS-CoV-2
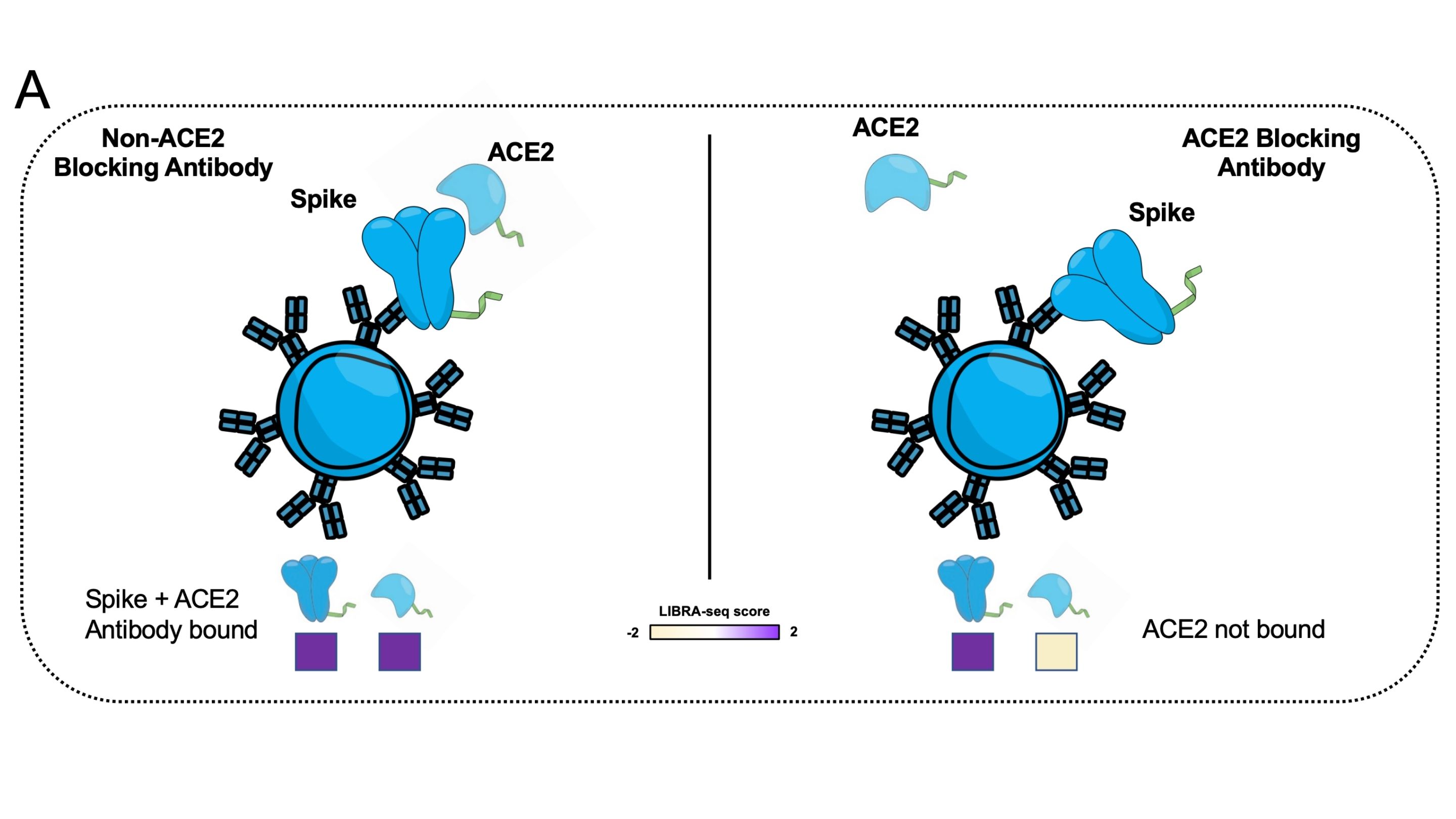
Dr. Georgiev’s group has recently upgraded LIBRA-seq via adding a new modality: ligand blocking(4). This smart move can rapidly identify the ultra-potent neutralizing antibodies that both target the antigen and also block the binding of antigen to its native ligand. For example, the entry of SARS-CoV-2 virus into cells is mediated by binding of viral spike protein and host cell surface receptor ACE2. Thus it is necessary to identify antibodies that simultaneously neutralize the SARS-CoV-2 virus and prevent its binding to host ACE2.
To achieve this, Dr. Georgiev’s group added human ACE2 protein conjugated with a unique DNA oligo barcode into traditional LIBRA-seq (DNA barcoded SARS-CoV-2 spike antigen library + B cells from COVID-19 convalescent donors with previous SARS-CoV2 infection) (Fig 4). After sequencing, they compute the LIBRA-seq score for both viral spike protein antigen and human ACE2. This allows for the identification of B cells that have high LIBRA-seq scores for the target antigen (strongly binds to spike protein) and low LIBRA-seq scores for the ligand (weakly binds to ACE2), as B cells that are likely able to potently block antigen-ligand interactions.
Recovering the antibody sequences from these B cells will facilitate rapid development of highly-effective therapeutic monoclonal antibodies. Using LIBRA-seq with ligand blocking, they identify an ultra potent SARS-CoV2 neutralizing antibody 5317-4 with high affinity for spike protein (IC50 value=7.3ng/mL) and significantly blocks ACE2 binding to spike protein (4).
Only a few papers are published on LIBRA-seq and it will be very interesting to see additional papers and applications for this novel technique.







