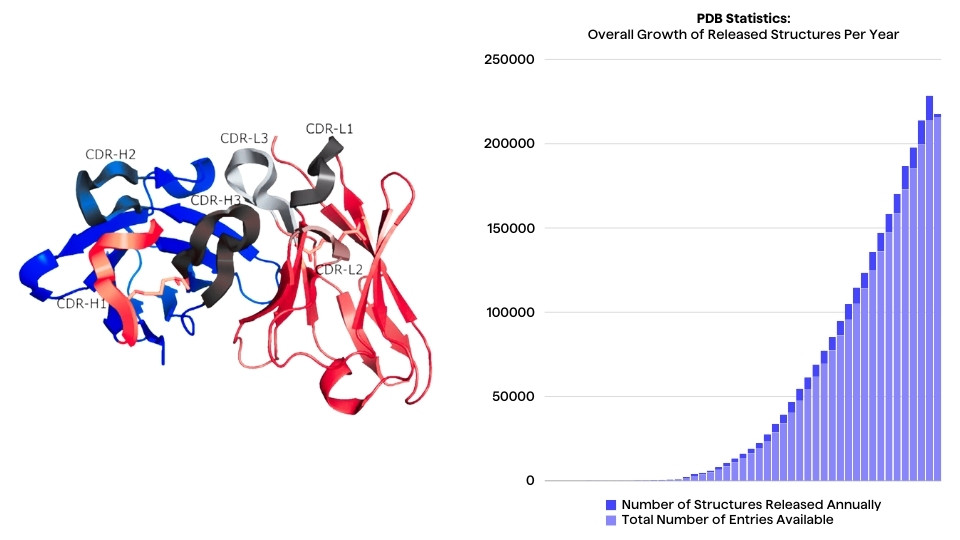
Engineered monoclonal antibodies (mAbs) play an increasingly important part in novel immunotherapeutics and to date, they constitute a large proportion of approved therapeutic antibodies.
The humanization of monoclonal antibodies involves conserving the IgG variable regions of the animal antibody and replacing the IgG constant region (Fc) with those from human immunoglobulins. Approved therapeutic antibodies include a majority of humanized full-length IgGs and other formats such as
- sdAb (single-domain antibody), e.g. envafolimab
- Fab (Fragment antigen-binding), e.g. certolizumab, idarucizumab and ranibizumab
- scFv (single-chain fragment variable), e.g. brolucizumab
- Bispecific antibodies, e.g. emicizumab, faricimab and amivantamab
With the plethora of innovative engineered antibody formats, a challenging aspect from an engineering and comparability perspective remains standard numbering and definitions of antibody regions. Accurate definitions of the crucial antigen-binding regions – the complementarity determining regions (CDRs) – is therefore paramount.
The strength of a specific paratope-epitope binding interaction depends on several characteristics of the binding sites, including the shape (CDR length, orientation) of the regions as well as biophysical properties such as the hydrophobicity and electrostatic forces of the interacting residues1. Framework regions (FRs) and the Fc may also affect the binding affinity or specificity of an antibody2.
While definitions of antibody variable regions may vary between different standard schemes, these definitions and numbering schemes serve as indispensable tools for properly distinguishing CDRs from FRs in antibody engineering and development.
Standard definitions of antibody variable regions
Different antibody numbering schemes have been developed for accurate identification and subsequent comparison of variable regions of different antibodies. The most commonly used numbering schemes include IMGT, Kabat, Chothia, Martin (Enhanced Chothia or AbM) and Honneger’s numbering scheme (AHo).
The largest differences between the numbering schemes arise from where insertion points of amino acids are placed in regions. These insertion points allow amino acids to be inserted or deleted which occur as a result of somatic hypermutation. Below we will briefly describe the most commonly used antibody numbering schemes.

Kabat
Building on earlier work in the 1970s, the Kabat numbering system (first published for immunoglobulins by Kabat et al. in 19793) was originally derived from observations from sequence alignments of light chain λ, κ and heavy chain sequences for a number of antibodies and for the α, β, γ, δ chain sequences for a number of TCRs.
The scheme defines specific positions where insertions and gaps may occur in CDRs and FRs. In the system, additional amino acid insertions are annotated with letters1.
As the original dataset was fairly limited and with standard-length variable regions, the numbering and definitions in Kabat are less flexible. Therefore, antibodies of unconventional length or with unconventional insertions or deletions may be overlooked when they do not match the more stringent regions defined in Kabat.
Another caveat with the Kabat numbering scheme is that it fails to take into account the topology or 3d structure of the binding domains.
IMGT
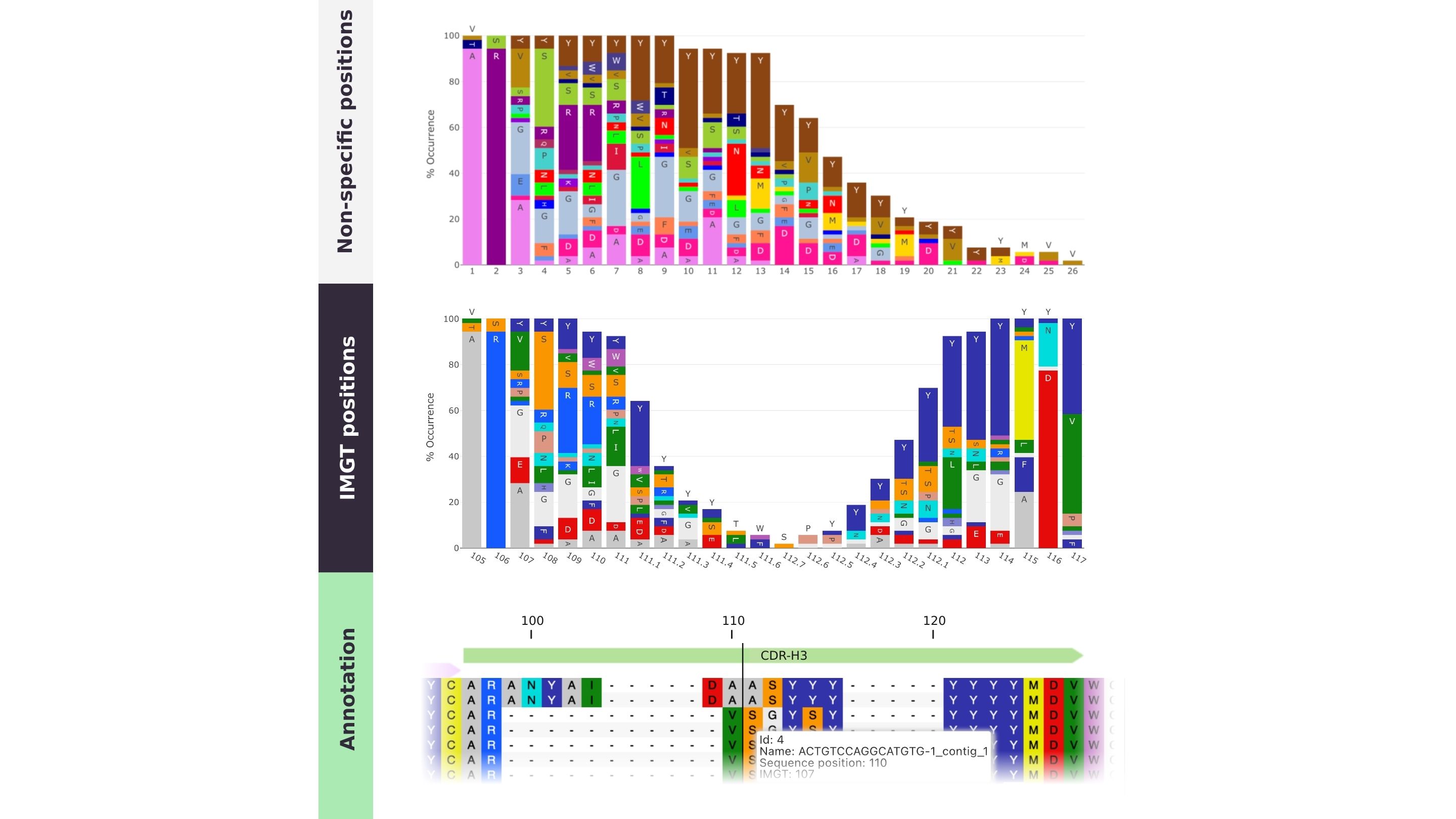
The IMGT numbering scheme was originally based on alignment of germ-line V genes, spanning from FR1 to the beginning of the CDR3.The scheme was later extended to cover the entire variable region. The numbering runs from 1 to 128 based on the V-gene sequence alignment, with an insertion point only between positions 111 and 112 in the CDR3 for lengths exceeding 13 amino acids.
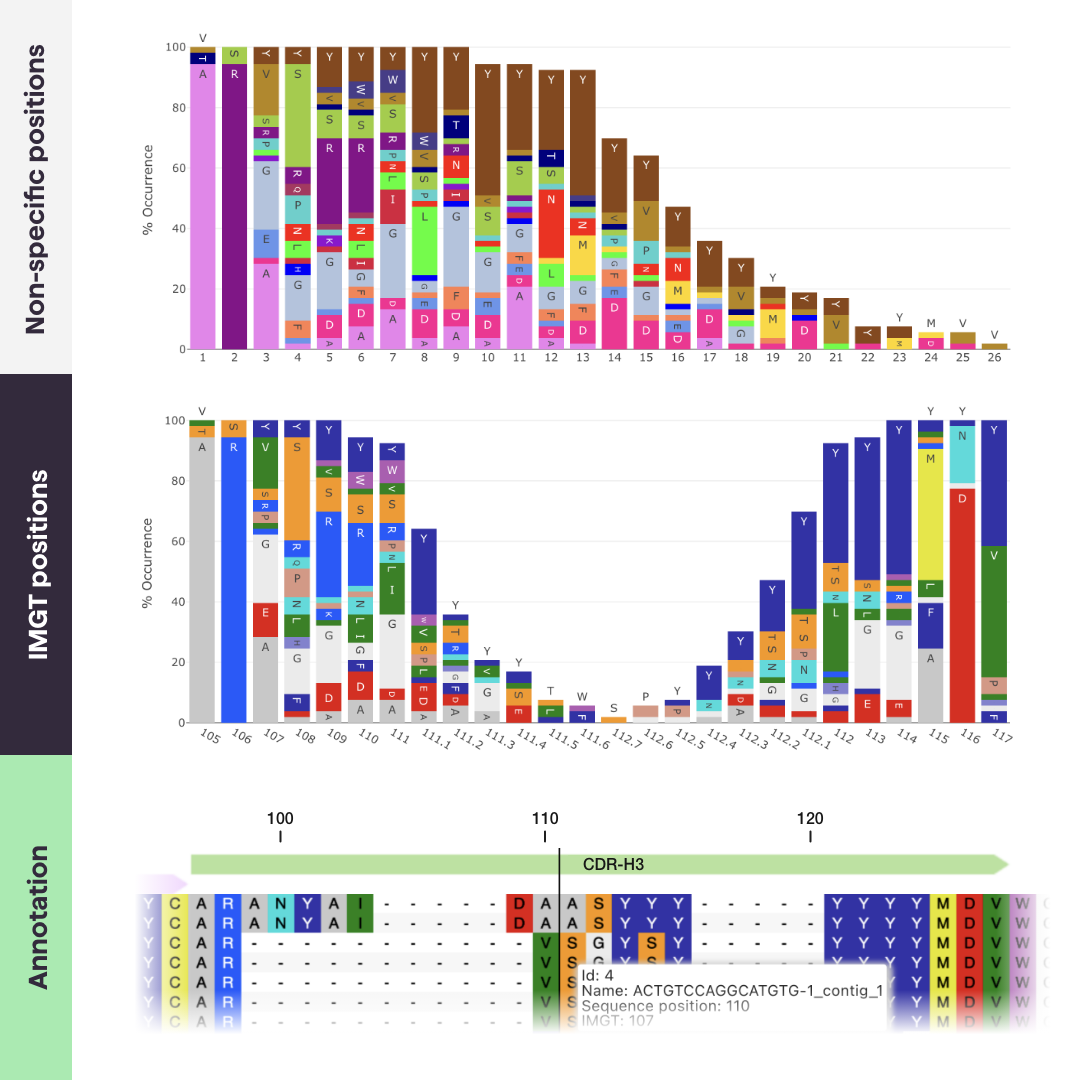
The sequence alignments behind the numbering scheme are based on a complete reference gene database, encompassing the entire immunoglobulin superfamily. This makes IMGT a widely applicable and widely applied standard scheme.
However, the flexibility and structural correlation of the scheme is limited when it comes to insertions of new amino acids, as they are appended to the CDRs.
Chothia
Chothia is a structure-based scheme, created (by Chothia and Lesk5) by aligning the variable region crystal structures forming CDRs instead of a sequence-based alignment. Differences between Kabat and Chothia can be found in amino acid insertion points, for example for CDR-L1 and CDR-H1, as well as the loop lengths in CDRs.
In short, Chothia numbering corresponds with the three-dimensional structures of hypervariable regions of typical length antibodies.
Martin
Martin is a scheme that is essentially an updated version of Chothia, including proposed corrections to certain positions, which are based on analysis of a database of both sequences and structures.
With greater sequence variability in the reference dataset, also unconventional CDR lengths and deletions are taken into account in the numbering scheme.
CDR definitions from sequences
Accurately predicting the loop-region and structure of the CDRs from amino acid sequences is an extremely useful tool.
Studies on the canonical classes have indeed indicated that CDR-L 1,2 and 3 as well as CDR-H1 and 2 have a more limited amount of conformations, with often preserved residues in certain positions5, 6 . However, regions outside the CDRs, e.g. the non-canonical “DE-loop”7, may also affect the ultimate structure of the loops.
Several computational tools have also been developed recently to predict loop structure from large amounts of sequences, including the ABlooper8, SCALOP9, AlphaFold210.
These deep learning-based tools have the potential to bring increasing insights for designing and optimizing antibody structures in antibody engineering and represent a new wave of computational research in the field.
Some open-source tools, such as ANARCI11, allow you to apply numbering to a given translated amino acid sequence with different numbering schemes.
PipeBio’s annotation tools allow annotation of DNA or protein sequences by aligning them to the closest reference germline gene sequence, adding definitions for FRs, CDRs the Fc-region, and applying a numbering scheme, such as IMGT, Kabat or AbM.
Conclusion
While standard antibody numbering schemes exist, deciding which numbering scheme to use depends on the premise of the research objective.
- Sequence alignment-based schemes (Kabat, IMGT) offer the benefit of large reference databases and
- A large number of derivative tools that can be used with the schemes to accurately define a wide array of antibody regions.
- IMGT is widely applicable scheme for standard numbering of antibodies and TCRs, regardless of the chains or species.
- Numbering systems with CDR definitions that correspond to antibody CDR loop structure (Chothia and Martin) can be good alternatives for antibody engineering efforts where structure and the interacting residues are the focus.
In the end, the final objective and needs determine what type of a scheme is best for each use case, since using custom numbering and derivatives of the most widely used systems is also an option.


.jpg)