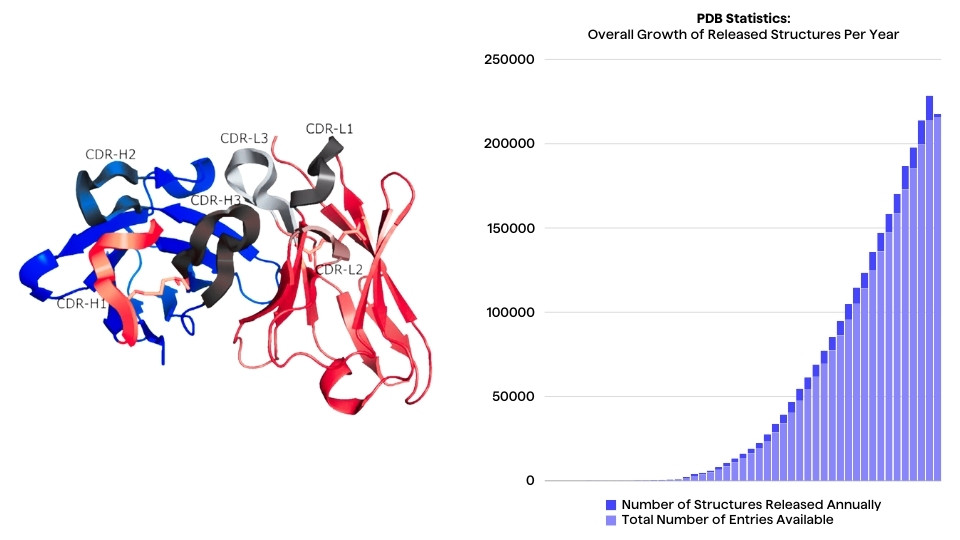
Biologics are becoming the drugs of choice when targeting cancers, infectious diseases etc. Monoclonal antibodies have taken up 6 out of 10 of the top selling drugs in the latter years and biologic drugs are expected to keep being some of the most efficient and potent drugs available.
To find good drug candidates new sequencing technologies are becoming an integral part of drug discovery campaigns as it has been in molecular biology labs and diagnostic labs since the initial discovery of the DNA helical structure by Watson and Crick in 1953 (Watson et al., 1953)
History
The most known sequencing methods developed in the mid-seventies were Maxam-Gilbert and Sanger sequencing. Maxam-Gilbert sequencing was developed in 1976 and made use of radioactive labeled phosphorus in the DNA backbone and not routinely used anymore. Frederik Sanger developed Sanger Sequencing, which quickly became popular and has been widely adopted. It is still heavily used in laboratories today.
In the late nineties new and massively scalable sequencing technologies started to emerge. These were commonly known as “Next generation sequencing” or “High throughput sequencing” methods and the most known companies at the time were Solexa and 454. Solexa was acquired by Illumina and 454 technologies acquired by Roche. Numerous other companies have worked to revolutionize the sequencing technologies and the in addition to Illumina are the major (current) companies Oxford Nanopore and PacBio (Pacific Biosystems). A more thorough review of NGS platforms is described by Hodkinson et al.
Sequencing and antibody drug discovery
Traditionally, antibodies have been sequenced with "Sanger sequencing" technology and is still today highly used because of superior quality and read length. The throughput of sanger sequencing is low, thus next generation sequencing is often a desired approach for sequencing antibody libraries or repertoires. The shorter read length and higher error rates can often be accommodated by the higher throughput of NGS.
With the increased amounts of sequencing data, scientists are faced with new challenges in bioinformatics data analysis. With Sanger sequencing, it is possible to handle and analyse the sequence data manually, but with NGS this is no longer possible. PipeBio has developed a unique bioinformatics analysis platform which enables the end user to analyse massive amounts of NGS data themselves freeing up bioinformaticians for more specialised work.
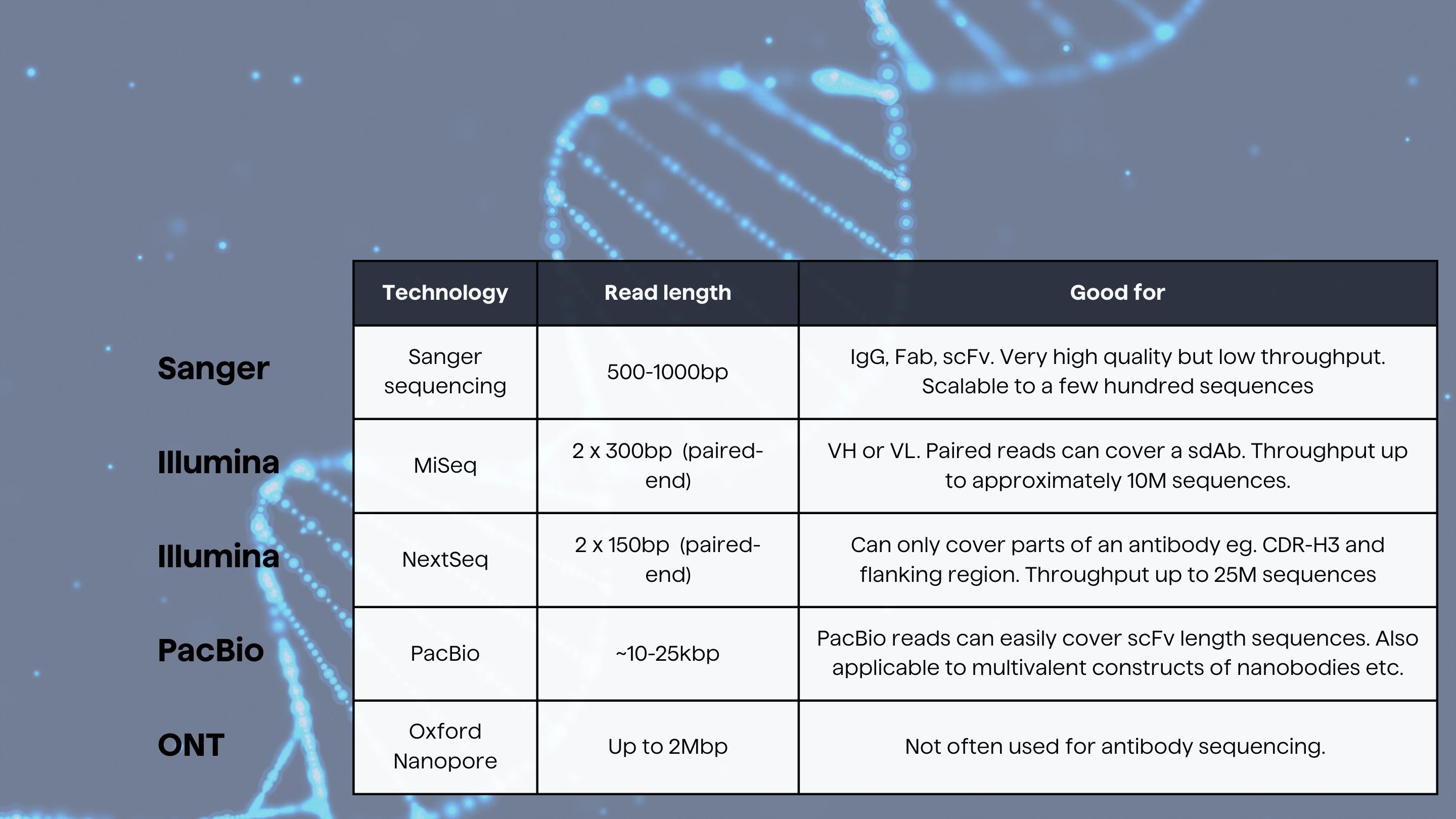
We here summarize some of the most used sequencing technologies used in antibody drug discovery.
Sanger sequencing
Sanger sequencing has been routinely used in labs for more than 20 years and will produce very high-quality results, although the throughput is low. Sanger sequencing has very often been used when sequencing antibody expressing hybridomas, single domain antibodies, scFv etc.

Illumina
Illumina is a major player in the sequencing market and has developed several unique instruments of which some are good for antibody sequencing.
The downside of the Illumina sequencing is the relatively short read lengths which are to a certain extent covered by the ability to do pair-end sequencing. Typically, pair-end libraries of 2x250bp or 2x300bp can be used to cover the entire variable domain of an antibody which is typically around 400-500bp. It is possible to use MiSeq’s for sequencing for example scFv but that will typically require a more advanced lab protocol as well as bespoke bioinformatics analysis.
Illumina has several instruments with various sequence output volumes and read lengths which can be found on their website.

NGS is often used for sequencing Phage Display libraries or repertoire sequencing or in conjunction with single cell platforms such as 10x Genomics.
PacBio
PacBio has developed a long-read sequencing technology which is capable of sequencing single molecules. The long reads produced by PacBio makes it a good choice for sequencing scFv’s, Fabs and similar which cannot be easily covered by paired end Illumina reads. PacBio has notoriously been known for poor sequencing quality but recent versions of their instrument together with advanced bioinformatics processing of the raw data has significantly improved sequence quality.

Oxford Nanopore
Oxford Nanopore has a versatile sequencing platform which has not yet been widely used in antibody discovery. Singh et al. published a paper where they made use of nanopore sequencing on single cell level.

Summary
Brief summary of major sequencing technologies used for antibody drug discovery. Significant deviations from this can be found and will depend on library construction and laboratory work and this should not be used as a guide to select which technology to use.
PipeBio Bioinformatics Cloud supports Sanger, NGS and PacBio sequence data and we have a wide range of tool for building custom analysis pipelines for your needs.


.jpg)