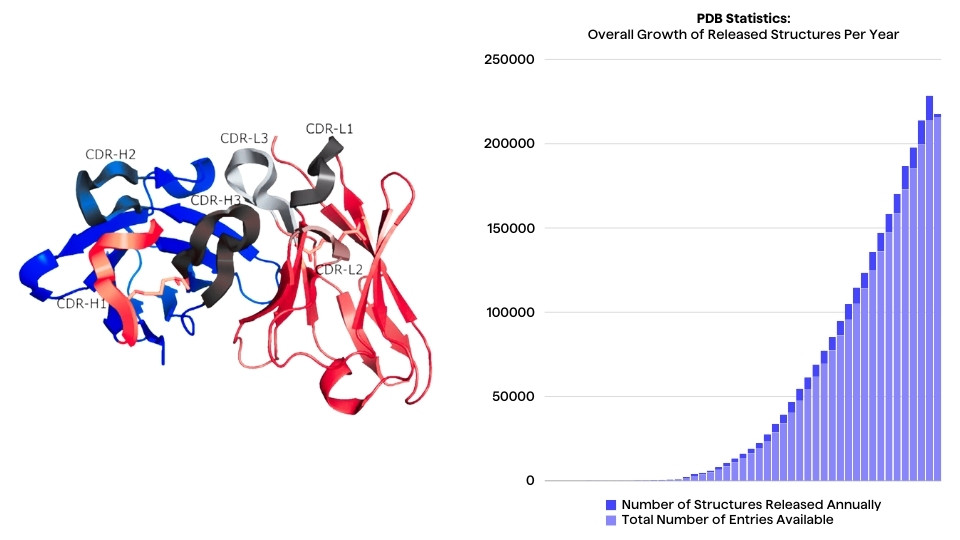
Research for discovery and development of in silico designed therapeutic antibodies has accelerated with the surge of data, computational resources and research into, among others, natural language-based machine learning (ML) models.
The models adapted from natural language processing (NLP) have turned out to translate well for identifying patterns in the language that we use in biology as well, making them a promising tool for inferring complex dependencies in DNA and amino acid sequences.
Today, there are numerous ML strategies out there each with a distinct choice of architecture, dataset design, data encoding.
In terms of ML architectures, some commonly used ones include
- Recurrent neural networks (RNN)
- Convolutional neural networks (CNN)
- Generative adversarial networks (GAN)
- Transformers
Generally, encodings of antibody sequences or sequence information can be done through one-hot-encoding with a binary 0/1 value representing each amino acid in a particular position. Substitution matrices such as the BLOSUM matrix, scores or distance matrices can also be used to reflect particular amino acid properties or three-dimensional structures.
The dataset design aspect is crucial and includes the dimensionality of the data and which factors, e.g. epitope-paratope structure, binding affinity and binding specificity, are considered.
Various in silico and in vitro approaches are also combined for specific antibody discovery research purposes.
Combined computational and experimental approaches
New methods that leverage advances in computational processing power, high-throughput data generation methods, novel algorithms and computational models are becoming increasingly prominent in cutting-edge research.
Established in vitro approaches such as display technologies (including phage, yeast and mammalian cell) used for antibody discovery and engineering are being augmented and optimized with computational approaches.
Fully computational approaches, such as de novo sequence design of antibodies are also being developed, with the first-ever computationally designed antibody, AU-007 by Aulos Bioscience (designed by Biolojic Design), having reached clinical trials.1
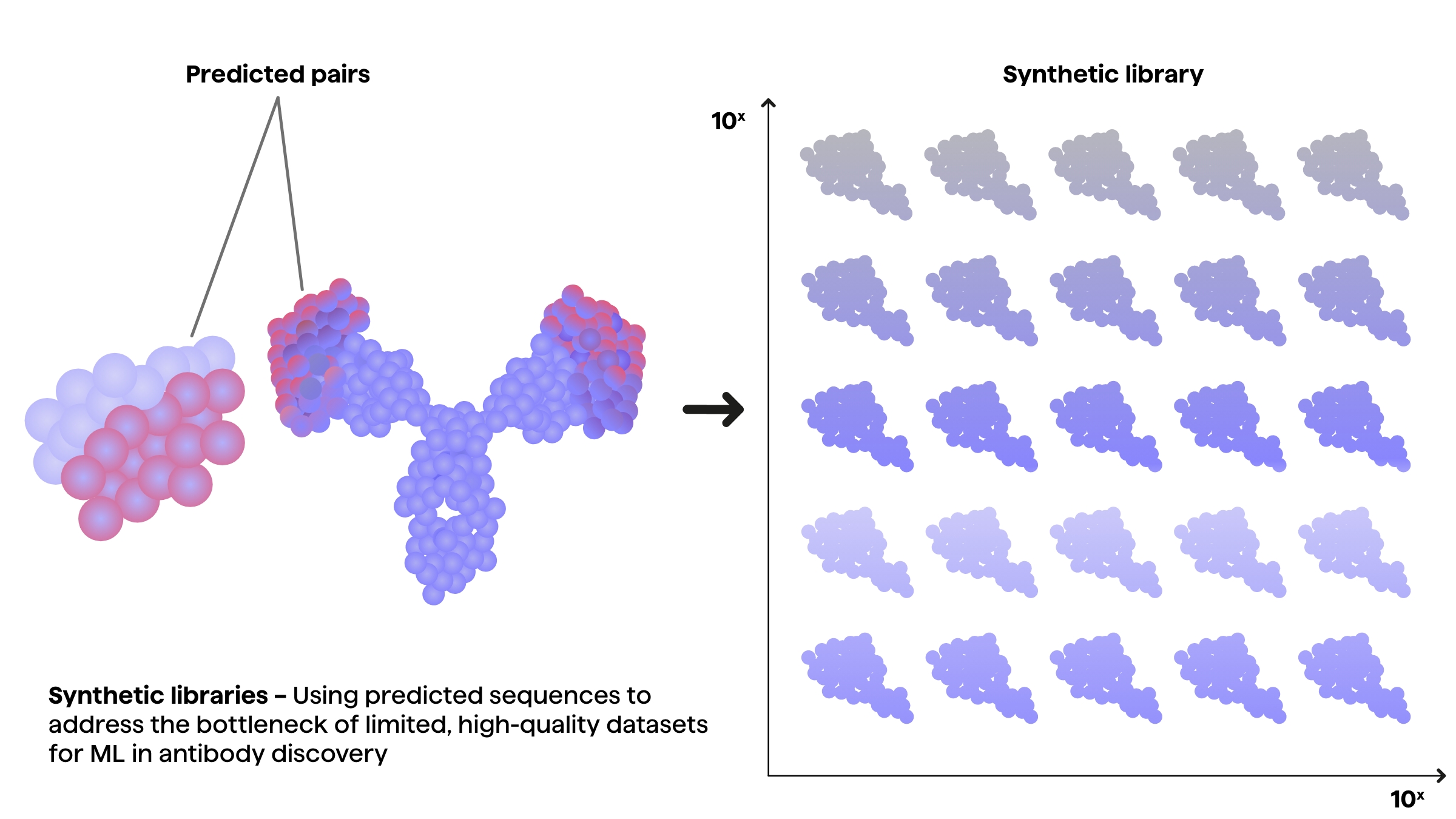
ML-assisted antibody engineering approaches can be used for creation of in silico libraries through prediction or directed mutagenesis to explore previously inaccessible combinatorial spaces of antibody sequences. Identifying candidate sequences from synthetic libraries can yield previously unseen antibodies with higher affinity, specificity and better biophysical properties.
The synthetic libraries can also be used as further input for models that optimize candidate sequences or even de novo design of novel therapeutic molecules.
Examples of machine learning approaches
Improving affinity through prediction
To name a few examples of ML approaches, Saka et al. (2021) show in their paper how affinity maturation of antibodies against the metabolite kynurenine could be improved by prediction of higher affinity sequences.
A long short term memory network (LSTM) ML model was trained on a set of enriched sequences after biopanning and used to predict the affinity of virtual sequences and select the most promising candidates.

Impressively, the affinity of generated sequences was found to be over 1800-fold higher than that of the parental clone.2
Co-optimization of site-specific and non-specific binding
Makowski et al. (2022)3 designed a model for co-optimizing site-specific and non-specific binding of emibetuzumab, with the aim to predict continuous features (as opposed to binary) from binary classifications. First, a library of pre-predicted sequences with low non-specificity was generated through site-mutation of CDRs.
Subsequently, the library was displayed in yeast cells, sorted for affinity and specificity, deep sequenced and selected for frequency. Finally, both a relatively simple linear discriminant analysis (LDA) model and neural network models were created to predict antibody affinity and specificity (with similar model performance).
A lead candidate was then chosen to be further optimized based on additional representational features of the VH domains.
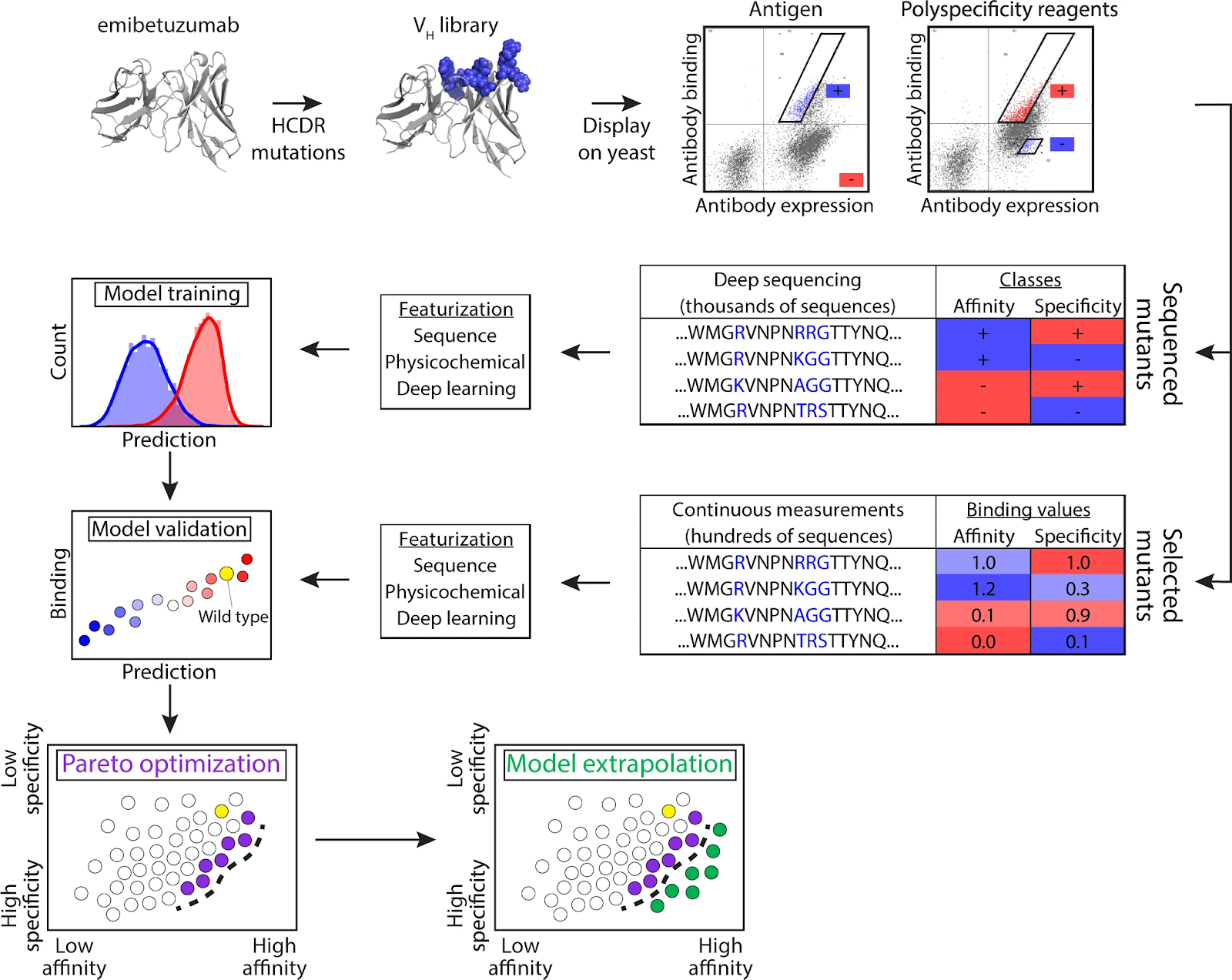
Predicting high-specificity variant binders
In another paper from 2021, Mason et al.5 describe a deep learning approach for predicting high-specificity variant binders of trastuzumab (Herceptin).
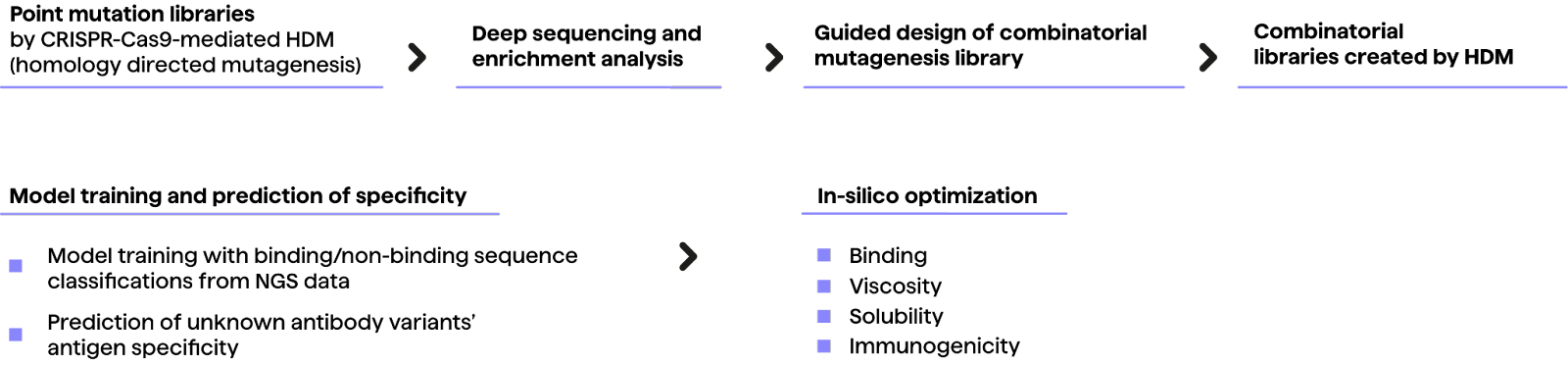
The authors induced mutations to the CDR-H3 regions of a non-binding hybridoma cell line to create point-mutated trastuzumab variants, deep sequenced for enrichment, and screened for specificity to HER2 by cell sorting (FACS).
The screening was performed to determine which amino acid positions were likely to impact specificity and direct generation of a combinatorial library. Binding/non-binding data from the initial library was then used to create the combinatorial library with a theoretical sequence space of 7.17 x 108 variants.
The original non-binding cell line was subsequently used to express sequences from the combinatorial library in hybridoma cells. Approximately 4 x 104 binders and non-binders were then identified across three rounds of enrichment through deep sequencing and used to train a convolutional neural network (CNN) model.
The CNN model was then used to classify sequences from a library of 7.2 x 107 sequences based on predicted specificity. The result was 6.96 x 106 predicted binders. Experimental validation of an unfiltered sample showed retained specificity for HER2.
Ultimately, a subset of the predicted binders were further optimized for properties including viscosity, clearance, solubility and immunogenicity by in silico, sequence-based filtering methods. In conclusion, the study showcases how antibody discovery can be accelerated by predictive models.
In silico approaches and multidimensional data
Predicting sequence dependencies and especially sequence-function dependencies, for example antibody-antigen interactions, relies heavily on the generation of high quality, structured training data.
This data may include information about the specific conformational structures of the antibody’s paratope and antigen’s epitope, affinity and specificity.
The complexity of a given binding event, the conditions affecting it as well as limited empirical data on existing paratope-epitope pairs pose a challenge for computational methods accurately predicting such interactions.
For instance, combined information about paratope, epitope and binding affinity is particularly scarce and datasets limited. While more multidimensional data is constantly being generated from experiments, it might still fall short of what ML models would require as input for accurate predictions.
In recent years, several machine learning methods have been developed to provide structural models of proteins (AlphaFold27), CDR loop structures (ABlooper8, SCALOP9) and antibody-antigen 3D structure libraries6.

The 3D structure of CDRs is especially of interest when predicting antibody-antigen complementarity. However, optimizing for one factor might come at a cost of increased immunogenicity or undesired pharmacokinetic effects.
Predictive models might be able to assist with the creation of synthetic libraries that in turn can be used as input for future ML models.
Synthetic datasets from ML predictions
As mentioned, the size and quality of datasets used for ML models has come to play a crucial part in enabling predictive models to predict antibody interactions, structures and sequences.
Although vast amounts of sequencing data and increasing amounts of binding property data for paratope-epitope pairs is constantly created, synthetic datasets are often needed for ML models.

The purpose of synthetic datasets is to both act as a benchmark for predictions and enable the models to be applied to smaller experimental datasets.
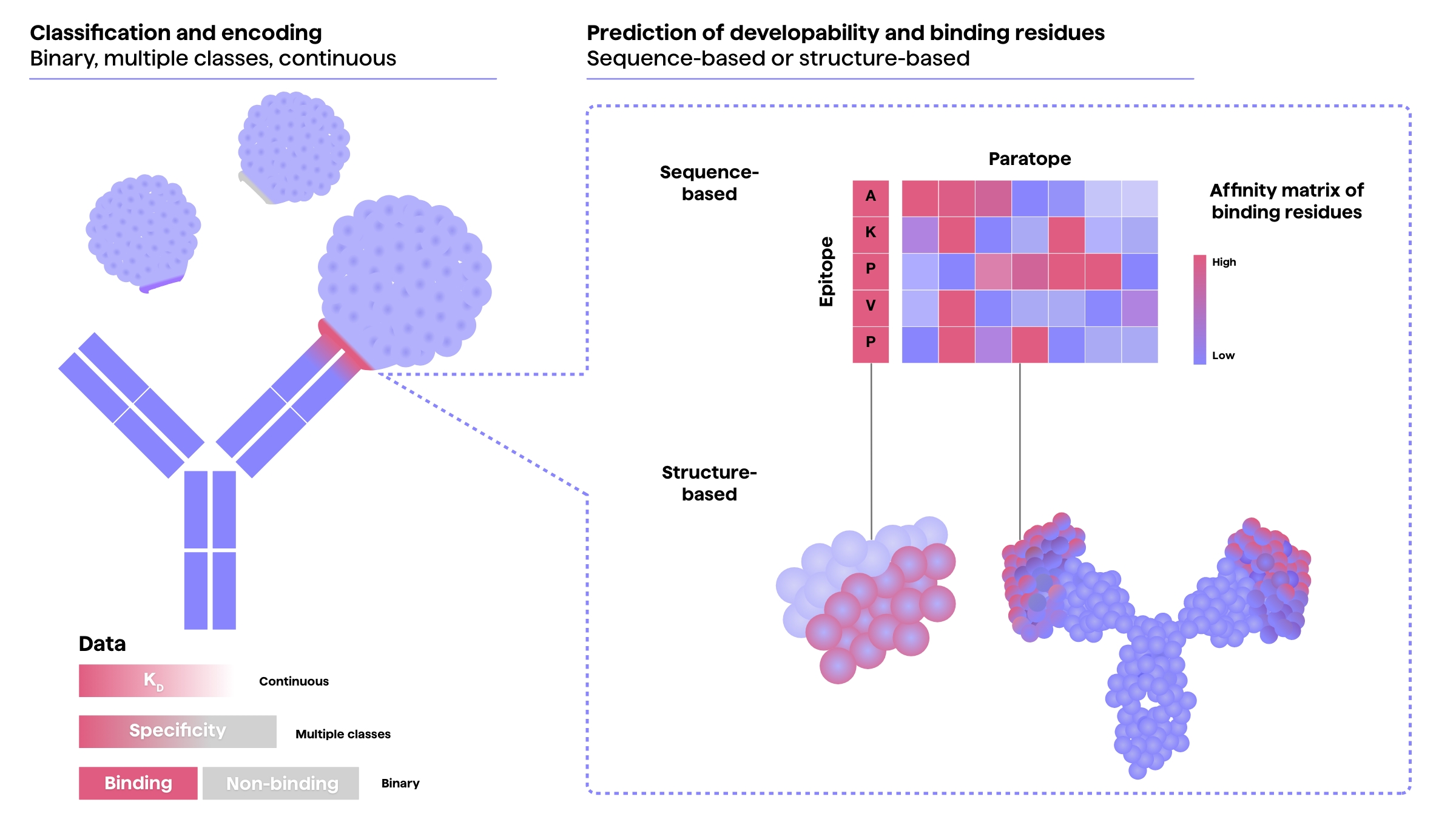
Akbar et al. (2022)11 list a few machine learning tasks that most ML approaches adopt with regard to antibody-antigen interactions – a crucial aspect of antibody therapeutics.
These include:
- Classification of the binding properties (e.g. binding/not binding, affinity and specificity),
- Predicting antibody sequence developability or affinity to target, as well as
- Predicting the binding amino acid residues for a given paratope-epitope pair.
Data encoding approaches to these can be sequence-based, structure-based or both structure and sequence-based.
Predicted library of CDR3-antigen pairs
In their paper, Akbar et al. (2022) describe the generation of an impressive 6.9 million synthetic binding structures of murine CDR-H3 regions with 159 antigens.
This resulted in around 1 billion antibody-antigen pairs with conformational epitope-paratope structures and their affinities. The library was created to model the biological complexity of the structures to a computationally feasible extent.
Called ‘Absolut!’, the library was evaluated with machine learning models trained on Absolut!-predicted structures or experimental datasets. Prediction accuracy of the models were shown to be similar both when trained on the synthetic dataset and experimental ones.
The study shows how by creating increasingly large synthetic libraries and using them as training data for machine learning models can help alleviate the data bottlenecks currently constraining the predictive power of ML models in antibody therapeutics.
Conclusion
In conclusion, advances in generation of predicted and experimentally verified multidimensional data remain fairly application-specific to date.
One seemingly certain thing seems to be that aggregation of experimental assay data and antibody sequence data in a curated and labeled format will be essential for enabling the data-driven approaches for biologics research in upcoming years.
The choice of ML model, architecture and data encoding and design remain fairly application-specific but are continuously being applied to new approaches in antibody discovery.


.jpg)